The Liberty16
Stay ahead of the curve with the mobile, real-time system PCR that fits in the palm of your hand
Smaller
A truly hand-held qPCR device
Mobile
Operates for up to three hours on re-chargeable battery power
Cloud-enabled
Bluetooth connectivity to a native iPhone app for access to test results
Fast
Gold-standard results in less than 20 minutes

Easier and more accessible testing for SARS-CoV-2 (COVID-19)
Ubiquitome has partnered with the Yale School of Public Health to enhance public access to COVID-19 testing, with United States Food and Drug Administration (FDA) emergency use authorization (EUA) of the combination of SalivaDirect™ with Ubiquitome’s Liberty16 mobile real time-PCR.
Supporting faster testing for SARS-CoV-2
Isothermal amplification of the SARS-CoV-2 gene via LAMP technology with positive results in as little as 10 mins.
Publications
Explore the following publications to discover how our PCR device is revolutionizing the way we understand, diagnose, and combat diseases.
Abstract
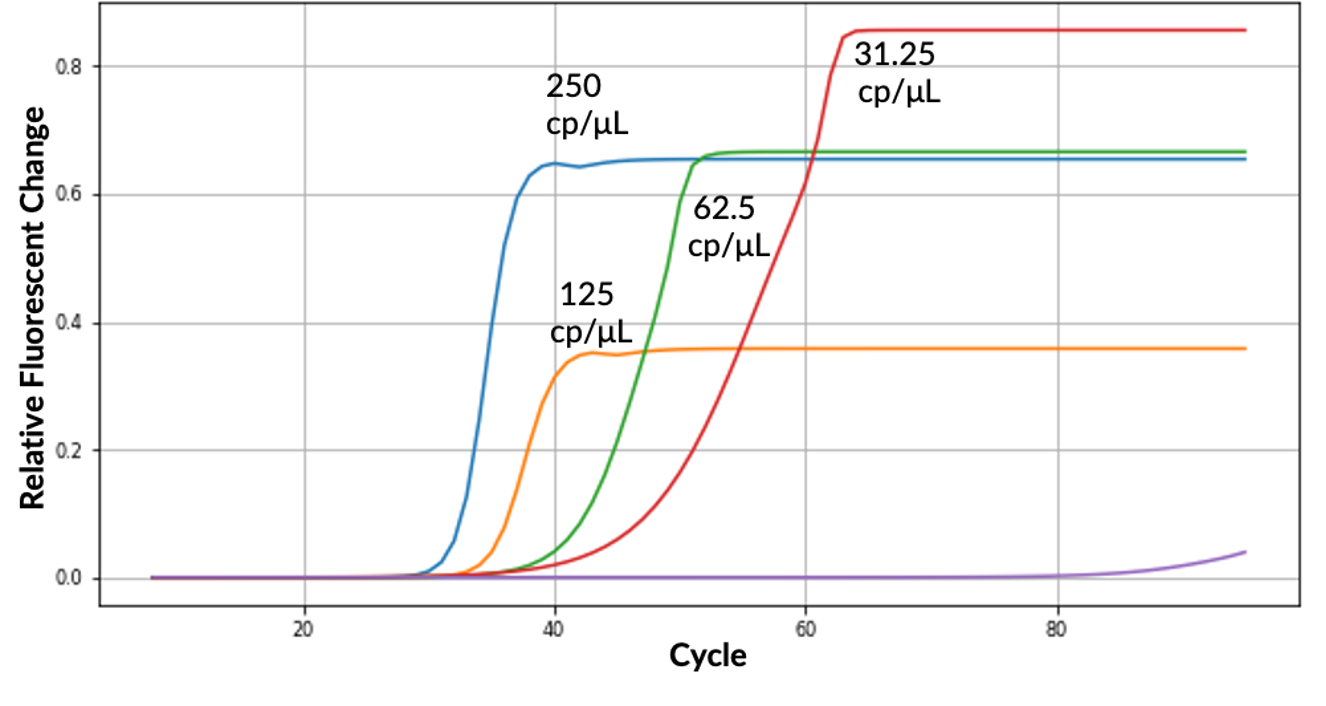
The COVID-19 pandemic has highlighted the need and benefits for all communities to be permitted timely access to on-demand screening for infectious respiratory diseases. This can be achieved with simplified testing approaches and affordable access to core resources. While RT-qPCR-based tests remain the gold standard for SARS-CoV-2 detection due to their high sensitivity, implementation of testing requires high upfront costs to obtain the necessary instrumentation. This is particularly restrictive in low-resource settings. The Ubiquitome Liberty16 system was developed as an inexpensive, portable, battery-operated single-channel RT-qPCR device with an associated iPhone app to simplify assay set-up and data reporting. When coupled with the SalivaDirect protocol for testing saliva samples for SARS-CoV-2, the Liberty16 device yielded a limit of detection (LOD) of 12 SARS-CoV-2 RNA copies/µL, comparable to the upper end of the LOD range for the standard SalivaDirect protocol when performed on larger RT-qPCR instruments. While further optimization may deliver even greater sensitivity and assay speed, findings from this study indicate that small portable devices such as the Liberty16 can deliver reliable results and provide the opportunity to further increase access to gold standard SARS-CoV-2 testing.
Keywords: COVID-19; PCR; SARS-CoV-2; diagnostics; saliva.
Abstract
A significant limitation of quantitative real-time polymerase chain reaction (qPCR) technology for environmental research has been the lack of portability of the amplification and detection equipment needed to perform qPCR assays in the field. The ability to perform qPCR assays in the field would significantly improve the utility of the technology, enabling quicker risk management decisions in acute environmental emergencies, and speeding the pace of environmental research. We tested a portable qPCR instrument, Liberty16, to evaluate if this instrument would be useful for qPCR analysis in our fieldwork. Parallel testing of the Liberty16 with our existing LightCycler 480 Instrument II revealed similar repeatability and sensitivity for the detection of targets in known and blind freshwater samples by qPCR. Both instruments were able to detect all target sequences at 20 copies per reaction and up to 66% targets in a 10-fold dilution. In seven of eight blind freshwater samples, results from both instruments were consistent for detected or nondetected target. In the other sample, the target was only detected by the Liberty16. Portable instruments of this type may open up new avenues for researchers to perform qPCR analyses in the field.
Abstract
African swine fever (ASF) is a contagious viral hemorrhagic disease that affects domestic pigs and wild boar. The disease is notifiable to the World Organization of Animal Health (WOAH), and causes significant deaths and economic losses. There is currently no fully licensed vaccine available. As a result, early identification of the causative agent, ASF virus (ASFV), is crucial for the implementation of control measures. PCR and real-time PCR are the WOAH-recommended standard methods for the direct detection of ASFV. However, under special field conditions or in simple or remote field laboratories, there may be no sophisticated equipment or even stable electricity available. Under these circumstances, point-of-care systems can be put in place. Along these lines, a previously published, rapid, reliable, and electricity-free extraction method (TripleE) was used to isolate viral nucleic acid from diagnostic specimens. With this tool, nucleic acid extraction from up to eight diagnostic samples can be realized in one run in less than 10 min. In addition, the possibility of completely omitting viral DNA extraction was analyzed with so-called direct real-time PCR protocols using ASFV original samples diluted to 1:40 in RNase-free water. Furthermore, three real-time PCR cyclers, developed for use under field conditions (IndiField, Liberty16 and UF-300 GenecheckerTM), were comparatively applied for the sensitive high-speed detection of ASFV genomes, with overall PCR run times between 20 and 54 min. Depending on the viral DNA extraction/releasing method used and the point-of-care cycler applied, a total time for detection of 30 to 60 min for up to eight samples was feasible. As expected, the limitations in analytical sensitivity were positively correlated to the analysis time. These limitations are acceptable for ASFV diagnostics due to the expected high ASFV genome loads in diseased animals or carcasses.
Keywords: African swine fever virus; DNA isolation; point-of-care (POC); portable real-time PCR.
Abstract
To assist the global eradication of peste des petits ruminants virus (PPRV), a molecular test for the rapid and reliable detection of PPRV was developed which additionally enables the detection of pathogens relevant for differential diagnostics. For this purpose, the necessary time frame of a magnetic bead-based nucleic acid extraction protocol was markedly shortened to 7 min and 13 s. The optimized extraction was run on a BioSprint 15 platform. Furthermore, a high-speed multi-well RT-qPCR for the genome detection of PPRV and additional important pathogens such as Foot-and-mouth disease virus, Parapoxvirus ovis, Goatpox virus, and Mycoplasmacapricolumsubsp.capripneumoniae was established and combined with suitable internal control assays. The here-described qPCR is based on a lyophilized master mix and takes only around 30 to 40 min. Several qPCR cyclers were evaluated regarding their suitability for fast-cycling approaches and for their diagnostic performance in a high-speed RT-qPCR. The final evaluation was conducted on the BioRad CFX96 and also on a portable Liberty16 qPCR cycler. The new molecular test designated as "FastCheckFLI PPR-like", which is based on rapid nucleic acid extraction and high-speed RT-qPCR, delivered reliable results in less than one hour, allowing its use also in a pen-side scenario.
Keywords: Small ruminant morbilli virus; differential diagnosis; fast extraction; high-speed RT-qPCR; molecular pen-side test; peste des petits ruminants virus (PPRV); rapid detection method.
The Liberty16
Our devices are designed and manufactured under an ISO 13485:2016 certified Quality Management System.

| Block Format | 16 wells |
| Thermal system | Peltier-based system |
| Consumables | Industry standard 8 strip PCR tube and caps |
| Sample volume | 10μl to 40μl |
| Average ramp rate | 2.3°C/sec |
| Ambient operation | 10°C – 40°C |
| Calibrated dyes | SYBR Green, FAM |
| Data collection | Data collected for all wells |
| Battery | Internal - rechargeable lithium ion |
| DIMENSIONS | |
| Outside dimensions | Length closed: 21.2 cm (8.35 inches) Width: 11 cm (4.33 inches) Height: 11.5 cm (4.53 inches) |
| Outside dimensions | 3.2 kg (7.05 lb) |
